Paul Fitzpatrick, Department of Biochemistry and Structural Biology
University of Texas at San Antonio
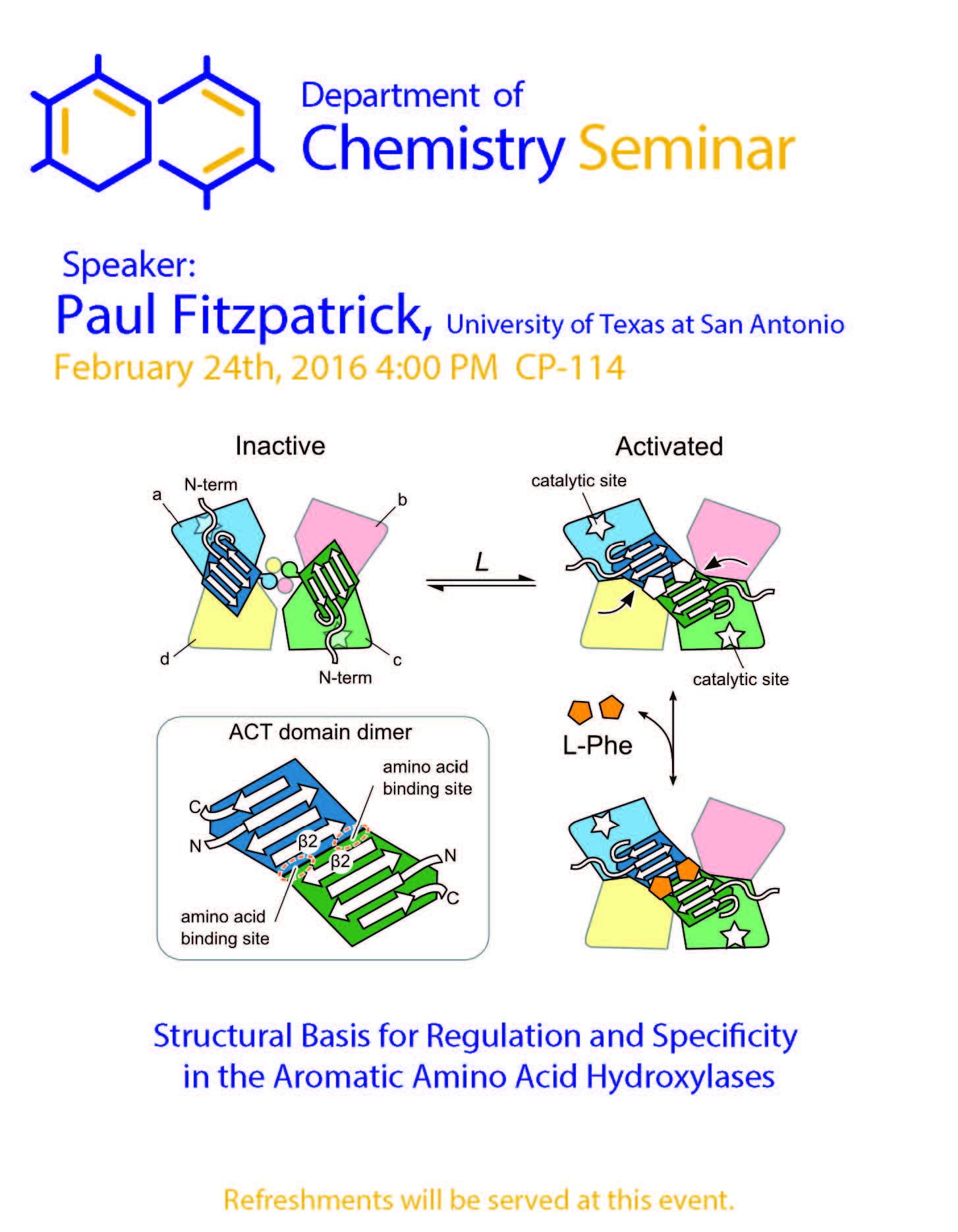
Structural Basis for Regulation and Specificity in the Aromatic Amino Acid Hydroxylases
The non-heme iron-containing enzymes tyrosine hydroxylase and phenylalanine hydroxylase catalyze the hydroxylation of the aromatic side chains of their respective substrates. Phenylalanine hydroxylase initiates the catabolism of excess phenylalanine in the diet; a deficiency in the enzyme results in the disease phenylketonuria. Tyrosine hydroxylase catalyzes the rate-limiting step in the biosynthesis of the catecholamine neurotransmitters dopamine, norepinephrine, and epinephrine. The catalytic domains of the two enzymes have very similar structures, and detailed studies of the catalytic mechanisms have established that both enzyme utilize the same mechanism. Mutagenesis of Asp425 in tyrosine hydroxylase converts the enzyme to a highly efficient phenylalanine hydroxylase. Saturation mutagenesis of this residue has provided insight into the basis for this change in specificity.
The sequences of their N-terminal regulatory domains differ significantly, consistent with divergent regulatory mechanisms. Tyrosine hydroxylase is regulated by a balance between feedback inhibition and phosphorylation, while phenylalanine hydroxylase is allosterically regulated by phenylalanine. Using a combination of structural approaches we have determined the structural basis for the regulation of the two enzymes, showing that a common structural fold underlies discrete regulatory mechanisms.