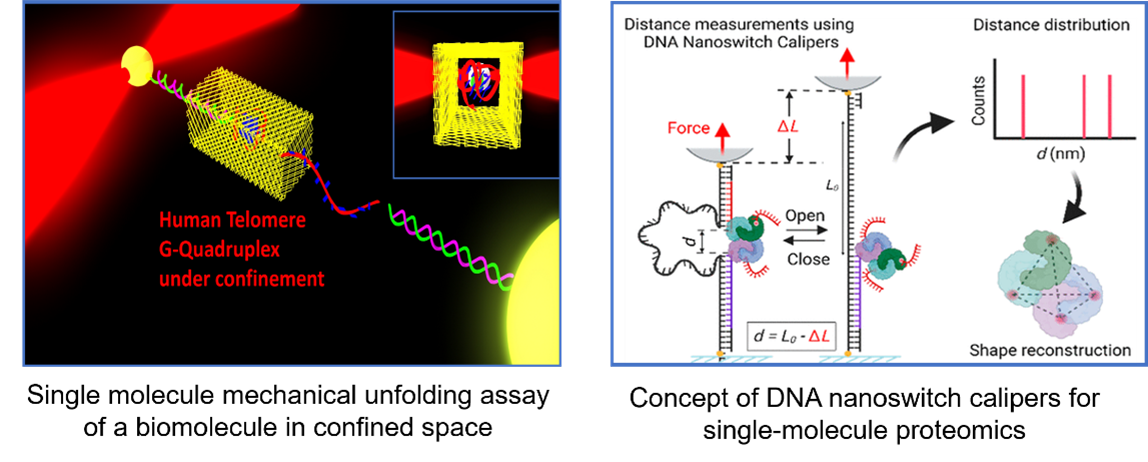
Abstract
The accurate identification and detailed analysis of biomolecules have led to a deeper understanding of biological intricacies, paving the way for innovative therapeutic strategies. The cutting-edge field of single-molecule techniques has emerged as a highly promising avenue in this pursuit of revealing the identity and real-time dynamics of biomolecular structure and interactions. In this seminar, I will discuss the development of single-molecule bioanalytical approaches, from unraveling the stability and dynamics of folded nucleic acid structures to proteomics applications. By employing DNA nanotechnology techniques to create a confined space for a G-quadruplex (GQ) structure and performing single-molecule mechanical unfolding assay of GQ using optical tweezers, we revealed that confined space facilitates the folding of the G-quadruplex structure by enhancing both stability and kinetics. Venturing into single-molecule proteomics, we introduced the mechanically reconfigurable DNA Nanoswitch Calipers (DNC) capable of measuring multiple coordinates on single biomolecules with angstrom-level precision. By measuring the distances of specific amino acid residues in optical and multiplexed magnetic tweezers, our work extends to the single molecule fingerprinting of peptides, showcasing discrimination within a heterogeneous population and even between distinct post-translational modifications. Furthermore, by using force-activated barcodes in measuring the distances of biotin-binding sites in single native-folded biotin-streptavidin complexes, we demonstrated the DNC’s potential in single-molecule structural proteomics applications.