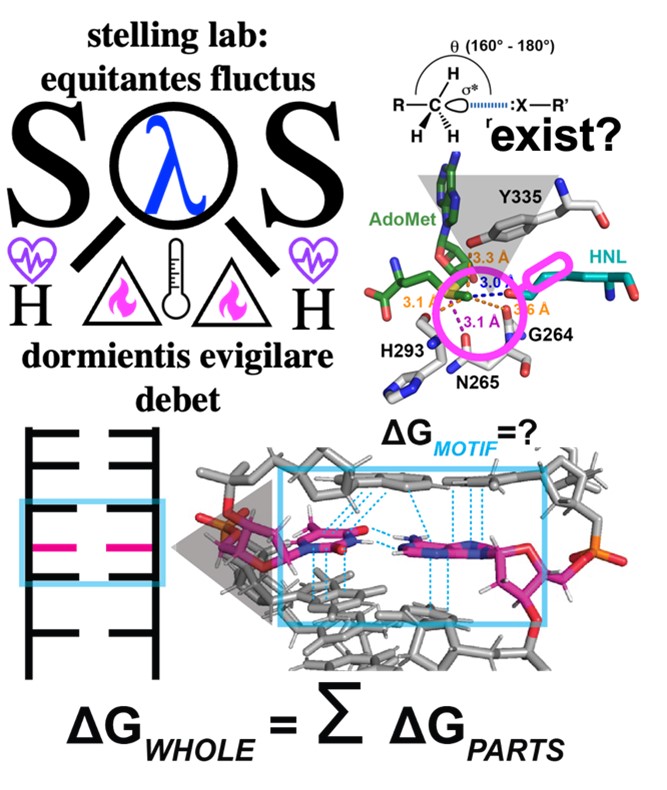
 Adoption of the correct three dimensional structures by biomolecular complexes in solution is essential for their performance in cells, and the formation of individual non-covalent interactions can be critical to their stability and functions. There is thus much interest in developing solution-state methods of detection to determine the arrangement of atoms at particular locations in these complexes, and that are sensitive to the stabilities of the local structural motifs they form. The Stelling group develops the vibrational states of nucleic acids and an essential substrate for a broad range of enzymatic reactions, s-adenosyl-l-methionine (AdoMet), as solution state sensors of local structure and energetics when they are contained within the large number of complexes that bind them in cells. Over the short term, the dimensions of specific vibrational states of the nucleic acids and AdoMet will be determined via rigorous isotope labeling and computational studies. Over the long term, these submolecular-scale sensors will be deployed to examine local structure and dynamics in the active sites of the vast number of enzymes that use AdoMet as a substrate, and to probe nucelobase structure and stability in DNA and RNA containing nuceloprotein complexes.
Adoption of the correct three dimensional structures by biomolecular complexes in solution is essential for their performance in cells, and the formation of individual non-covalent interactions can be critical to their stability and functions. There is thus much interest in developing solution-state methods of detection to determine the arrangement of atoms at particular locations in these complexes, and that are sensitive to the stabilities of the local structural motifs they form. The Stelling group develops the vibrational states of nucleic acids and an essential substrate for a broad range of enzymatic reactions, s-adenosyl-l-methionine (AdoMet), as solution state sensors of local structure and energetics when they are contained within the large number of complexes that bind them in cells. Over the short term, the dimensions of specific vibrational states of the nucleic acids and AdoMet will be determined via rigorous isotope labeling and computational studies. Over the long term, these submolecular-scale sensors will be deployed to examine local structure and dynamics in the active sites of the vast number of enzymes that use AdoMet as a substrate, and to probe nucelobase structure and stability in DNA and RNA containing nuceloprotein complexes.
Applying atomic scale vibrational probes for solution state measurements of local structure and stability within complete biomolecular complexes
Date:
-
Location:
CP 114
